|
Ethnobotanical Leaflets 12: 609-13. 2008.
Analysis of Inter-Species Relationships of Ocimum Species Using RAPD Markers
*Harisaranraj R., **R. Prasitha, *S. Saravana Babu and *Suresh K.
*Department of Plant Biology and Plant Biotechnology, Chikkaiah Naicker College, Erode. (T.N.) INDIA **Vivekananda College of Arts and Science College for Women, Trichengode. (T.N.)
Issued 11 August 2008
Abstract Genetic inter-relationship of Seven Ocimum species was estimated using Random Amplified Polymorphic DNA (RAPD) markers. The 15 selected RAPD primers out of 2 primers were amplified in all Ocimum species. O. basilicum has very close similarity (89%) with O. tenuiflorum and another two species of O. gratissimum and O. micranthum. Our results suggested that genetic relationships in Ocimum species using RAPD banding data may be useful for plant improvement and an efficient way to conserve genetic resources of Ocimum species, in addition to their effective medicinal uses. Key Words: Lamiaceae, Ocimum, DNA, RAPD.
Introduction
The purpose of the present study was to use RAPD markers to evaluate the genetic relationships between seven Ocimum species. Ocimum belongs to the Lamiaceae family, which has close to 252 genera and 6700 species (Mabberley, 1997) most of which are used as medicine (Wren, 1968). The leaves are often hairy and posses epidermal glands which secrete volatile oils giving characteristic scents to many of the species. The essential oils found in leaves, seeds, flowers and roots of Ocimum species are used as medicine. Under in vitro conditions, the oils have shown to have antibacterial activity against gram positive bacteria: Staphylococcus aureus (ATCC 25923), Bacillus species and gram negative bacteria: Escherichia coli (ATCC 25922), Salmonella typhi, Pseudomonas aeruginosae, Proteus mirabilis, Klebsiella pneumoniae (ATCC 27853), Salmonella enteriditis, Shigella flexineri, and pathogenic fungus namely Candida albicans (Nakamura et al., 1999; Matasyoh et al., 2007). Because of its potential as a traditional medicine, incorporation of O. species into agro forestry systems would not only make the species accessible to the majority of the rural population that uses it but also contribute to its genetic conservation. However, before widespread domestication of the species is implemented, it would be important to determine its genetic diversity in India so that useful genotypes that could be used as cultivars by farmers can be selected thereby also facilitating the efficient conservation, management and utilization of the species genetic diversity. There are techniques available for assessing genetic diversity at molecular level. Those are morphology, biochemical and more recent DNA technology. The application of DNA technology in agricultural research has progressed rapidly over the last twenty years, especially in the area of cultivar identification and characterization (Nybom, 1990) as well as determination of population diversity in many plant species (Lei et al., 2006; Chen and Yang, 2004; Nan et al., 2003; Ipek and Madison, 2001; Muluvi et al., 1999; Cardoso et al., 2000). Recently global interest in oriental medicine, production of those plants has grown even more over the following years. Since many species and varieties exist, development of molecular markers would be important for quality assessment in the medicinal industry (Sang-Bok Lee, et al, 2000). During the last decade several novel DNA-markers (RAPD, RFLP, SSR, ISSR etc.) have been rapidly integrated into the tools available for genome analysis. Salimath et al., (1995) has been used for DNA fingerprinting and assessing genetic diversity. Presence or absence of DNA bands in the gel may be used as RAPD markers to study close genetic relationship (Sang-Bok Lee, et al., 2000), inter- and intra-specific genetic variations (M’Ribu and Hilu 1994), for the identification of specific genes (Paran et al. 1991; Martin et al. 1991), and to study the pattern of gene expression (Valle et al. 2000).
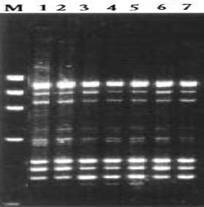
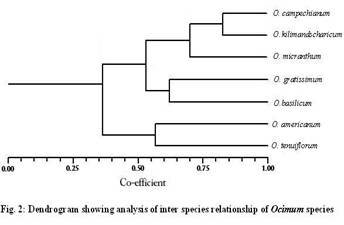
Materials and Methods Genetic material Young leaves of seven Ocimum species viz., O. tenuiflorum, O. americanum, O.basilicum, O. gratissimum, O. micranthum, O. kilimandscharicum ,O. campechianum were collected from Southern India and preserved at – 80°C. DNA extraction Stored leaves were pulverized in liquid nitrogen and DNA was extracted from each plant of Ocimum species according to the method described by Doyle and Doyle (1987). Total DNA was quantified spectrophotometrically and samples yielding good quality (A260/A280 ratio 1.7–1.9) and as well as visually by ethidium bromide staining on 0.8% agarose gel electrophoresis. RAPD Marker analysis A set of random 15 primers was purchased from a commercial source (Sigma Aldrich, Bangalore.). After initial tests, 15 primers (OPA-08; OPA-10; OPA-11; OPA-12; OPA-15; OPB-03; OPC-01; OPC-15; OPE-02; OPE-06; OPE-07; OPE-10; OPE-20; OPF-13; and OPF-20) were used for further studies. PCR reactions were performed according to the protocol of Williams et al. (1990). Briefly, DNA samples of the 7 Ocimum species of each individuals plants were adjusted to 20 ng/µl and used in the amplification reactions with a final volume of 25µl containing 1µl of DNA, 2 µl of primer (40 µM), 1 µl of dNTPs (10 mM), 0.2 µl Taq DNA polymerase (5 U/µl), 3 µl PCR buffer, 1.5 µl of MgCl2 (25 mM) and 16.3 µl dionized water. DNA amplification was carried out using Corbett Research thermocycler programmed with 3 min at 94°C for initial denaturation, followed by 35 cycles of 54sec at 94°C, 45 sec at 43°C, 2 min at 72°C, and a final 5 min extension at 72°C. After amplification, the DNA fragments were separated by electrophoresis for about 1 hour under constant voltage (50 V) in 1.5% agarose gel submersed in 1X TBE buffer. All PCR experiments were done at least twice and the best gels of the replicates were used for band scoring. The gels were stained with ethidium bromide solution and observed under ultraviolet light. Each gel was photo documented using the image capturing system bioprint. A 1 kb fragment size marker was used as a reference to allow comparison among the different gels (Genei 1kb ladder). Statistical analysis The amplified bands were scored as 1 and 0 based on band (allele) presence and absence, respectively. Sizes of amplified bands were estimated using Gel Pro analyzer software. The binary data set was used to calculate the pairwise Jaccard similarity index and to assemble the corresponding similarity matrix. The matrix obtained was used to generate a dendrogram using the UPGMA method (Unweigthed Pair Group Method Arithmetical Means). The distances in the dendrogram were compared with the genetic distances between genotype pairs to calculate the cophenetic correlation. All the analyses were performed with the aid of the 2006 version of the Non linear Dynamics -pc computer program. Results and Discussion Fifteen primers were used in this study of RAPD marker analysis to standardization of suitable specific primers amplifying the genetic materials of Ocimum species. All primers but two (OPE-20, and OPF-13) yielded maximum amplification products with all Ocimum species. Primers OPE-20 and OPF-13 amplifying all Ocimum species were produced many bands overall ranging in size 800 - 2800bp. The primers amplified DNA products from each Ocimum species generating reproducible band patterns. Primers OPE-20, and OPF- 13 generated uncommon band with DNA from some of the specimens, along with several common bands with each specimen .Primer OPE-20 was amplified in O. tenuiflorum very close similarity (89%) with O.basilicum, O. gratissimum and O. micranthum. Several polymorphic bands were detected. The remaining primers gave patterns that were identical or had differences too small to provide information on the genetic diversity. They could hardly distinguish the seven Ocimum species slightly different all Ocimum species. The PCR amplified band pattern of seven species of Ocimum species were shown in Figure.1 and the phylogenetic tree comprising a total of 7 Ocimum species RAPD marker was constructed as shown in Figure.2. The correlation coefficient calculated between RAPD when using the similarity 89% when using the dendrograms.
Fig. 1. PCR amplified band pattern of seven Ocimum species. 1. O. tenuiflorum 2.O. americanum 3. O.basilicum 4.O. gratissimum 5. O. micranthum 6.O. kilimandscharicum 7. O. campechianum
Ocimum species are valued as spice plants in India. Driven by commercial incentives, the wild population of this plant has been threatened with depletion in recent years due to excessive harvesting. The present study was preliminary attempt to develop RAPD primers to distinguish the seven Ocimum species showed that a more difficult screening of primers has to be done before RAPD markers can be developed. (Chatti et al. 2003) work done on the genetic diversity and the phylogenetic relation between 17 ecotypes analyzed by using the technique of RAPD marker and on nine species of Cinnamomum species by (Priya joy et al, 2008). This study showed a significant morphological variation and a large genetic diversity within and among cultivars. Lately, this technique has been used to study the genetic relations between the different species of coffee and to determine the relationship between hybrids (Paulo, 2003). The dendrogram supports the suggestion that similar variation of O.basilicum and O. gratissimum and also may be synonymous with other two species of O. tenuiflorum and O. micranthum. However the variation in terms of length 800-2800 bp and nucleotide the frequency of insertions and deletions is too large for phylogenetic analyses.
References
|